Journal IF and Current issue
Online first
Archive
About the Journal
Editorial Office
Editorial Board
Copy right and self-archiving policy
Peer review process
Instructions for Reviewers
Printed version subscription
Abstracting and indexing
Contact
Instructions for Authors
Policies
General information
Open Access, Licensing terms, Commercial use and Copyright terms policies
Self-archiving policy and Archive policies
Article Correction and Withdrawal policy
Manuscript Submission policy
Authorship policy
Conflict of Interest policy
Language considerations policy
Plagiarism and Duplicate publications policy
Ethics policy
Review process policy
Acceptance of manuscripts policy
Online First Articles and Special Issues policies
Generative artificial intelligence (AI) policy
Advertising policy
Article publication charges
Policies
General information
Open Access, Licensing terms, Commercial use and Copyright terms policies
Self-archiving policy and Archive policies
Article Correction and Withdrawal policy
Manuscript Submission policy
Authorship policy
Conflict of Interest policy
Language considerations policy
Plagiarism and Duplicate publications policy
Ethics policy
Review process policy
Acceptance of manuscripts policy
Online First Articles and Special Issues policies
Generative artificial intelligence (AI) policy
Advertising policy
ORIGINAL PAPER
Comprehensive proteomic analysis of the main liver
and attached liver of Glyptosternum maculatum on the basis
of data-independent mass spectrometry acquisition
1
Anhui Polytechnic University, Academy of Biological and Food Engineering, Department of Food Science and Engineering,
241000 Wuhu, China
2
Tibet Academy of Agricultural and Animal Husbandry Sciences, Academy of Aquatic Sciences, 850002 Lhasa, China
3
Tibet Academy of Agricultural and Animal Husbandry Sciences, Institute of Fisheries Science, 850002 Lhasa, China
4
Hefei Normal University, Academy of Life Science, Department of Biotechnology, 230601 Hefei, China
Publication date: 2022-10-25
J. Anim. Feed Sci. 2023;32(1):85-118
KEYWORDS
TOPICS
ABSTRACT
Glyptosternum maculatum Regan is a teleost species endemic to
the Brahmaputra River. This species has two livers, namely the attached liver
(AL), and the main liver (ML). However, the biological functions and molecular
mechanisms of these two livers remain uncharacterised. In this study, we used
a comparative quantitative proteomics involving data-independent acquisition
to elucidate the functions of ML and AL. A total of 63 differentially expressed
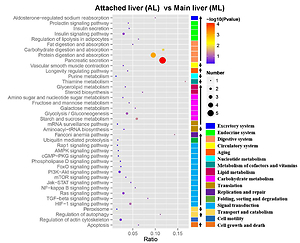
proteins (DEPs) were identified. According to the Gene Ontology enrichment
analysis, these proteins are involved in cellular processes, metabolic
processes, and hydrolase activities. Analysis of the Kyoto Encyclopedia of
Genes and Genomes indicated that the enriched pathways among these
DEPs were “pancreatic secretion” and “protein digestion and absorption” (i.e.,
digestive functions). Moreover, 569 highly expressed proteins in both ML and
AL were functionally annotated, revealing their diverse biological activities
that may involve adaptive responses to cold and hypoxic conditions at high
altitudes.
FUNDING
This work was supported by the Key Research and Development Projects in Tibet (No.
ZH20200002), Key Research and Development
Projects in Anhui (No. 201904e01020008), Key
Research and Development Projects in Tibet (No.
XZ202001ZY0040N), Anhui Province Foundation
for Talent in Higher Education (No. jxbjZD22),
Anhui Polytechnic University Scientific Research
Staring Foundation for the Introduction of Talent
(No. 2018YQQ011), Anhui Polytechnic University
Young and Middle-aged Talents Project (No. 2018-
2019), High Level Talent Introduction Project (No.
2018PTJB03), Talent Introduction Project of Hefei
32 Proteomic investigation of main liver and attached liver liver
Normal University (No. 403-60420050) and Qinghai Provincial Key Laboratory of Physical Geography and Environmental Process, Qinghai Provincial
Innovation Platform Construction Special Project
(No. 2020-ZJ-Y06).
CONFLICT OF INTEREST
The Authors declare that there is no conflict
of interest.
REFERENCES (62)
1.
Aberle L., Kruger A., Reber J.M. et al., 2020. PARP1 catalytic variants reveal branching and chain length-specific functions of poly (ADP-ribose) in cellular physiology and stress response. Nucleic Acids Res. 48, 10015–10033, https://doi.org/10.1093/nar/gk...
2.
Agrawal S., Karcher D., Ruf S., Bock R., 2020. The functions of chloroplast glutamyl-trna in translation and tetrapyrrole biosynthesis. Plant Physiol. 183, 263–276, https://doi.org/10.1104/pp.20....
3.
Akiyama M., Mizokami T., Miyamoto S., Ikeda Y., 2022. Kaempferol increases intracellular ATP content in C2C12 myotubes under hypoxic conditions by suppressing the HIF-1α stabilization and/or by enhancing the mitochondrial complex IV activity. J. Nutr. Biochem. 103, 108949, https://doi.org/10.1016/j.jnut...
4.
Altinok-Yipel F., Yipel M., Altug N., Ozdemir N., 2022. Blood concentrations of potentially toxic trace elements (PTEs) and correlation with biochemical and hematological parameters in dogs from thrace region, Turkey. Chemosphere 293, 133649, https://doi.org/10.1016/j.chem...
5.
Anderson L.L., Mao X., Scott B.A., Crowder C.M., 2009. Survival from hypoxia in C. elegans by inactivation of aminoacyl-tRNA synthetases. Science 323, 630–633, https://doi.org/10.1126/scienc...
6.
Anjo S.I., Santa C., Manadas B., 2017. SWATH‐MS as a tool for biomarker discovery: from basic research to clinical applications. Proteomics 17, 1600278, https://doi.org/10.1002/pmic.2...
7.
Beall C.M., 2007. Two routes to functional adaptation: Tibetan and Andean high-altitude natives. Proc. Natl. Acad. Sci. U.S.A. 104, 8655–8660, https://doi.org/10.1073/pnas.0...
8.
Birrell J.H., Shah A.A., Hotaling S., Giersch J.J., Williamson C.E., Jacobsen D., Woods H.A., 2020. Insects in high-elevation streams: life in extreme environments imperiled by climate change. Glob. Change Biol. 26, 1–18, https://doi.org/10.1111/gcb.15...
9.
Brose S.A., Marquardt A.L., Golovko M.Y., 2014. Fatty acid biosynthesis from glutamate and glutamine is specifically induced in neuronal cells under hypoxia. J. Neurochem. 129, 400–412, https://doi.org/10.1111/jnc.12...
10.
Bruderer R., Bernhardt O.M., Gandhi T. et al., 2015. Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated tree-dimensional liver microtissues. Mol. Cell. Proteomics 14, 1400–1410, https://doi.org/10.1074/mcp.M1...
11.
Castello P.R., David P.S., McClure T., Crook Z., Poyton R.O., 2006. Mitochondrial cytochrome oxidase produces nitric oxide under hypoxic conditions: implications for oxygen sensing and hypoxic signaling in eukaryotes. Cell Metab. 3, 277–287, https://doi.org/10.1016/j.cmet...
12.
Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M., 2014. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526, https://doi.org/10.1074/mcp.M1...
13.
Cunha I., Mangas-Ramirez E., Guilhermino L., 2007. Effects of copper and cadmium on cholinesterase and glutathione S-transferase activities of two marine gastropods (Monodonta lineata and Nucella lapillus). Comp. Biochem. Physiol. C Toxicol. Pharmacol. 145, 648–657, https://doi.org/10.1016/j.cbpc...
14.
Davis R.W., Polasek L., Watson R., Fuson A., Williams T.M, Kanatous S.B., 2004. The diving paradox: new insights into the role of the dive response in air-breathing vertebrates. Comp. Biochem. Phys. A Mol. Integr. Physiol. 138, 263–268, https://doi.org/10.1016/j.cbpb...
15.
Dietrich M., Judycka S., Żarski D., Malinowska A., Świderska B., Palińska-Żarska K., Błażejewski M., Ciereszko A., 2021. Proteomic analysis of pikeperch seminal plasma provides novel insight into the testicular development of domesticated fish stocks. Animal 15, 100279, https://doi.org/10.1016/j.anim...
16.
Gerasimovskaya E.V., Tucker D.A., Stenmark K.R., 2005. Activation of phosphatidylinositol 3-kinase, Akt, and mammalian target of rapamycin is necessary for hypoxia-induced pulmonary artery adventitial fibroblast proliferation. J. Appl. Physiol, 98, 722–735, https://doi.org/10.1152/japplp...
17.
Goeminne L.J.E., Gevaert K., Clement L., 2018. Experimental design and data-analysis in label-free quantitative LC/MS proteomics: a tutorial with MSqRob. J. Proteomics 171, 23–36, https://doi.org/10.1016/j.jpro...
18.
Han X., Chen L., Hu Z., Chen L., Sun P., Wang Y., Liu Y., 2021. Identification of proteins related with pemetrexed resistance by iTRAQ and PRM-based comparative proteomic analysis and exploration of IGF2BP2 and FOLR1 functions in non-small cell lung cancer cells. J. Proteomics 237, 104122, https://doi.org/10.1016/j.jpro...
19.
Hashim I.F., Mokhtar A.M.A., 2021. Small Rho GTPases and their associated RhoGEFs mutations promote immunological defects in primary immunodeficiencies. Int. J. Biochem. Cell Biol. 137, 106034, https://doi.org/10.1016/j.bioc...
20.
He W., Batty-Stuart S., Lee J.E., Ohh M. 2021. HIF-1α hydroxyprolines modulate oxygen-dependent protein stability via single VHL interface with comparable effect on ubiquitination rate. J. Mol. Biol. 433, 167244, https://doi.org/10.1016/j.jmb....
21.
Huijuan Z., 2011. Genesis of liver in Glyptosternum maculatum and related bioadaptive studies, in Library of Huazhong Agricultural University. Huazhong Agricultural University. Wuhan, HB (China)
22.
Ikeda D., Koyama H., Mizusawa N., Kan-No N., Tan E., Asakawa S., Watabe S., 2017. Global gene expression analysis of the muscle tissues of medaka acclimated to low and high environmental temperatures. Comp. Biochem. Physiol. Part D. Genomics Proteomics 24, 19–28, https://doi.org/10.1016/j.cbd....
23.
Jones P., Binns D., Chang H.Y. et al.,2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240, https://doi.org/10.1093/bioinf...
24.
Kelstrup C.D., Bekker-Jensen D.B., Arrey T.N., Hogrebe A., Harder A., Olsen J.V., 2017. Performance evaluation of the Q exactive HF-X for shotgun proteomics. J. Proteome Res. 17, 727–738, https://doi.org/10.1021/acs.jp...
25.
Kelstrup C.D., Young C., Lavallee R., Nielsen M.L., Olsen J.V., 2012. Optimized fast and sensitive acquisition methods for shotgun proteomics on a quadrupole orbitrap mass spectrometer. J. Proteome. Res. 11, 3487–3497, https://doi.org/10.1021/pr3000...
26.
Khan Z., Michalopoulos G.K., Stolz D.B., 2006. Peroxisomal localization of hypoxia-inducible factors and hypoxia-inducible factor regulatory hydroxylases in primary rat hepatocytes exposed to hypoxia-reoxygenation. Am. J. Pathol. 169, 1251–1269, https://doi.org/10.2353/ajpath...
27.
Khramtsov P., Kalashnikova T., Bochkova M., Kropaneva M., Timganova S., Rayev M., 2021. Measuring the concentration of protein nanoparticles synthesized by desolvation method: comparison of Bradford assay, BCA assay, hydrolysis/UV spectroscopy and gravimetric analysis. Int. J. Pharm. 599, 120422, https://doi.org/10.1016/j.ijph...
28.
Li H., Xie X., Li D., Chai Y., Liu H., Fan Q., Zhu B., 2017. Exo-celiac Liever in Glyptosternum maculatum. Prog. Nat. Sci. 17, 1109–1113
29.
Li Y., Burridge C.P., Lv Y., Peng Z., 2021. Morphometric and population genomic evidence for species divergence in the Chimarrichthys fish complex of the Tibetan Plateau. Mol. Phylogenet. Evol. 159, 107117, https://doi.org/10.1016/j.ympe...
30.
Liao X., Pan Q., Tian X., Wu X., Zhao F., 2022. Proteomic analysis of the electron uptake pathway of Rhodopseudomonas palustris CGA009 under different cathodic potentials. Process Biochem. 115, 42–48, https://doi.org/10.1016/j.proc...
31.
Liu H., Liu Q., Chen Z. et al., 2018. Draft genome of Glyptosternon maculatum, an endemic fish from Tibet Plateau. Gigascience 7, giy104, https://doi.org/10.1093/gigasc...
32.
Ma X., Dai W., Kang J., Yang L., He S., 2016. Comprehensive transcriptome analysis of six catfish species from an altitude gradient reveals adaptive evolution in Tibetan fishes. G3-Genes Genom. Genet. 6, 141–148, https://doi.org/10.1534/g3.115...
33.
McKenna M.C., Stevenson J.H., Huang X., Hopkins I.B., 2000. Differential distribution of the enzymes glutamate dehydrogenase and aspartate aminotransferase in cortical synaptic mitochondria contributes to metabolic compartmentation in cortical synaptic terminals. Neurochem. Int. 37, 229–241, https://doi.org/10.1016/S0197-...
34.
Murray A.J., Montgomery H.E., Feelisch M., Grocott M.P.W., Martin D.S., 2018. Metabolic adjustment to high-altitude hypoxia: from genetic signals to physiological implications. Biochem. Soc. Trans. 46, 599–607, https://doi.org/10.1042/BST201...
35.
Nassereddine S., Habbal R., Kassogue Y., Kaltoum A.B.O., Farah K., Majda H., Rhizlane A.E., Nadifi S., Dehbi H., 2021. Analysis of the influence of glutathione S-transferase (GSTM1 and GSTT1) genes on the risk of essential hypertension. Ann. Hum. Biol. 48, 585–589, https://doi.org/10.1080/030144...
36.
Nemkov T., Sun K., Reisz J.A., Song A. et al., 2018. Hypoxia modulates the purine salvage pathway and decreases red blood cell and supernatant levels of hypoxanthine during refrigerated storage. Haematologica 103, 361–372, https://doi.org/10.3324/haemat...
37.
Pan S., Zhang T., Rong Z. et al., 2017. Population transcriptomes reveal synergistic responses of DNA polymorphism and RNA expression to extreme environments on the Qinghai-Tibetan Plateau in a predatory bird. Mol. Ecol. 26, 2993–3010, https://doi.org/10.1111/mec.14...
38.
Pankaj K., Sugadev R., Sarkar S., Singh S.B., 2016. A network-based analysis of proteins involved in hypoxia stress and identification of leader proteins. J. Proteomics Enzymol. 5, https://doi.org/10.4172/2470-1...
39.
Peng X., Dai Z., Wang X., 2020. Comparative proteomic analysis to probe into the differences in protein expression profiles and toxicity bases of Latrodectus tredecimguttatus spiderlings and adult spiders. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 232, 108762, https://doi.org/10.1016/j.cbpc...
40.
Provost E., Weier C.A., Leach S.D., 2013. Multiple ribosomal proteins are expressed at high levels in developing zebrafish endoderm and are required for normal exocrine pancreas development. Zebrafish 10, 161–169, https://doi.org/10.1089/zeb.20...
41.
Qi L., Chen Y., Shi K., Ma H., Wei S., Sha Z., 2021. Combining of transcriptomic and proteomic data to mine immune-related genes and proteins in the liver of cynoglossus semilaevis challenged with Vibrio anguillarum. Comp. Biochem. Physiol. Part D Genomics Proteomics 39, 100864, https://doi.org/10.1016/j.cbd....
42.
Saliu J.K., Bawa-Allah K.A., 2012. Toxicological effects of lead and zinc on the antioxidant enzyme activities of post juvenile Clarias gariepinus. Resour. Env. 2, 21–26, https://doi.org/10.5923/j.re.2...
43.
Saxena V., Orgill D., Kohane I., 2007. A set of genes previously implicated in the hypoxia response might be an important modulator in the rat ear tissue response to mechanical stretch. BMC Genomics 8, 430, http://www.biomedcentral.com/1...
44.
Shah A.N., Cadinu D., Henke R.M., Xin X., Dastidar R.G., Zhang L. 2011. Deletion of a subgroup of ribosome-related genes minimizes hypoxia-induced changes and confers hypoxia tolerance. Physiol. Genomics 43, 855–872, https://doi.org/10.1152/physio...
45.
Shao Y., Wellman T.L., Lounsbury K.M., Zhao F.Q., 2014. Differential regulation of GLUT1 and GLUT8 expression by hypoxia in mammary epithelial cells. Am. J. Physiol. Regul. Integr. Comp. Physiol. 307, 237–247, https://doi.org/10.1152/ajpreg...
46.
Sharma N.K., Sethy N.K., Bhargava K., 2013. Comparative proteome analysis reveals differential regulation of glycolytic and antioxidant enzymes in cortex and hippocampus exposed to short-term hypobaric hypoxia. J. Proteomics 79, 277–298, https://doi.org/10.1016/j.jpro...
47.
Tran Q., Lee H., Park J., Kim S.H., Park J., 2016. Targeting cancer metabolism-revisiting the Warburg effects. Toxicol. Res. 32, 177–193, https://doi.org/10.5487/TR.201...
48.
Usman M., Zhao S., Jeon B.H., Salama E.S., Li X., 2022. Microbial β-oxidation of synthetic long-chain fatty acids to improve lipid biomethanation. Water Res. 213, 118164, https://doi.org/10.1016/j.watr...
49.
Velasco-Martínez I.C., Hernández-Camacho C.J., Méndez-Rodríguez L.C., Zenteno-Savín T., 2016. Purine metabolism in response to hypoxic conditions associated with breath-hold diving and exercise in erythrocytes and plasma from bottlenose dolphins (Tursiops truncatus). Phys. A Mol. Integr. Physiol. 191, 196–201, https://doi.org/10.1016/j.cbpa...
50.
Wang Y.Y., Zhou Y., Fu H.C., Huang H.Z., Li N.Q., Jin R.M., Fu X.Z., Li N.Q., 2021. Transcriptomic and proteomic analyses of the immune mechanism in pathogenetic and resistant mandarin fish (Siniperca chuatsi) infected with ISKNV. Aquaculture 545, 737198, https://doi.org/10.1016/j.aqua...
51.
Whiffen L.K., Midgley D.J., Mcgee P.A., 2007. Polyphenolic compounds interfere with quantification of protein in soil extracts using the Bradford method. Soil Biol. Biochem. 39, 691–694, https://doi.org/10.1016/j.soil...
52.
Wiśniewski J., Zougman A., Nagaraj N., Mann M., 2009. Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362, https://doi.org/10.1038/nmeth....
53.
Wong Y.H., Zhang Y., Lun J., Qiu J.W., 2021. A proteomic analysis of skeletal tissue anomaly in the brain coral Platygyra carnosa. Mar. Pollut. Bull. 164, 111982, https://doi.org/10.1016/j.marp...
54.
Xiao S.J., Mou Z.B., Yang R.B., Fan D.D., Liu J.Q., Zou Y., Zhu S.L., Zou M., Zhou C.W., Liu H.P., 2021. Genome and population evolution and environmental adaptation of Glyptosternon maculatum on the Qinghai-Tibet Plateau. Zool. Res. 42, 502–513, https://doi.org/10.24272/j.iss...
55.
Yang W., Liu J., Hou L., Chen Q., Liu Y., 2021. Shikonin differentially regulates glucose metabolism via PKM2 and HIF1α to overcome apoptosis in a refractory HCC cell line. Life Sci. 265, 118796, https://doi.org/10.1016/j.lfs....
56.
Yoon J., Kim Y.H., Min J., 2017. Evaluation of in vitro function by subcellular distribution of lysosomal and peroxisomal protein in Saccharomyces cerevisiae. J. Nanosci. Nanotechnol. 17, 244–250, https://doi.org/10.1166/jnn.20...
57.
Yu M., He S., 2012. Phylogenetic relationships and estimation of divergence times among Sisoridae catfishes. Sci. China Life Sci. 55, 312–320, http://dx.doi.org/10.1007/s114...
58.
Zera K., Sweet R., Zastre J., 2016. Role of HIF-1α in the hypoxia inducible expression of the thiamine transporter, SLC19A3. Gene 595, 212–220, http://dx.doi.org/10.1016/j.ge...
59.
Zera K., Zastre J., 2017. Thiamine deficiency activates hypoxia inducible factor-1α to facilitate pro-apoptotic responses in mouse primary astrocytes. PloS ONE 12, e0186707, http://dx.doi.org/10.1371/jour...
60.
Zhang G.Q., Mou Z., Xue W.H., Liu H., 2021. Phosphorylated protein modification analysis on normal liver and exo‐celiac liver of Glyptosternon maculatum. J. Fish Biol. 99, 1696–1707, https://doi.org/10.1111/jfb.14...
61.
Zhang H., Zhang W., Huang S., Xu P., Cao Z., Chen M., Lin X., 2022. The potential role of plasma membrane proteins in response to Zn stress in rice roots based on iTRAQ and PRM under low Cd condition. J. Hazard. Mater. 429, 128324, https://doi.org/10.1016/j.jhaz...
62.
Zhou X., Zhou L., Ge X., Guo X., Han J., Zhang Y., Yang H., 2020. Quantitative proteomic analysis of porcine intestinal epithelial cells infected with porcine deltacoronavirus using iTRAQ-Coupled LC-MS/MS. J. Proteome Res. 19, 4470–4485, https://doi.org/10.1021/acs.jp...
CITATIONS (1):
1.
Label-free and TMT-labeled proteomics methods to compare differences on normal liver + extra-celiac liver of Glyptosternum maculatum
Wu Sun, Yingying Yan, Zafarullah Muhammad, Yufeng Guo, Guoqiang Zhang
Wu Sun, Yingying Yan, Zafarullah Muhammad, Yufeng Guo, Guoqiang Zhang
Share
RELATED ARTICLE
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.