Journal IF and Current issue
Online first
Archive
About the Journal
Editorial Office
Editorial Board
Copy right and self-archiving policy
Peer review process
Instructions for Reviewers
Printed version subscription
Abstracting and indexing
Contact
Instructions for Authors
Policies
General information
Open Access, Licensing terms, Commercial use and Copyright terms policies
Self-archiving policy and Archive policies
Article Correction and Withdrawal policy
Manuscript Submission policy
Authorship policy
Conflict of Interest policy
Language considerations policy
Plagiarism and Duplicate publications policy
Ethics policy
Review process policy
Acceptance of manuscripts policy
Online First Articles and Special Issues policies
Generative artificial intelligence (AI) policy
Advertising policy
Article publication charges
Policies
General information
Open Access, Licensing terms, Commercial use and Copyright terms policies
Self-archiving policy and Archive policies
Article Correction and Withdrawal policy
Manuscript Submission policy
Authorship policy
Conflict of Interest policy
Language considerations policy
Plagiarism and Duplicate publications policy
Ethics policy
Review process policy
Acceptance of manuscripts policy
Online First Articles and Special Issues policies
Generative artificial intelligence (AI) policy
Advertising policy
ORIGINAL PAPER
Diversity of lactic acid bacteria associated with raw yak
(Bos grunniens) milk produced in Pakistan
1
Pir Mehr Ali Shah Arid Agriculture University, Institute of Food and Nutritional Sciences, Shamsabad, Muree Road, Rawalpindi 46000, Punjab, Pakistan
2
Pir Mehr Ali Shah Arid Agriculture University, Institute of Soil Sciences
Shamsabad, Muree Road, Rawalpindi 46000, Punjab, Pakistan
3
National Agricultural Research Centre (NARC), Food Sciences Research Institute, Centre Park Road, Islamabad 44000, Pakistan
4
Pir Mehr Ali Shah Arid Agriculture University, Institute of Biochemistry and Biotechnology, Shamsabad, Muree، Road, Rawalpindi 46000, Punjab, Pakistan
Publication date: 2021-02-28
Corresponding author
M. S. Ibrahim
Pir Mehr Ali Shah Arid Agriculture University, Institute of Food and Nutritional Sciences, Shamsabad, Muree Road, Rawalpindi 46000, Punjab, Pakistan
Pir Mehr Ali Shah Arid Agriculture University, Institute of Food and Nutritional Sciences, Shamsabad, Muree Road, Rawalpindi 46000, Punjab, Pakistan
J. Anim. Feed Sci. 2021;30(1):42-51
KEYWORDS
TOPICS
ABSTRACT
Yak (Bos grunniens) milk and its associated products are being
used in high altitudes of Northern Pakistan. The aim of the study was to elucidate
the diversity of lactic acid bacteria (LAB) isolated from raw yak milk. In total, 88
isolates were characterized according to their phenotypic, physiological and
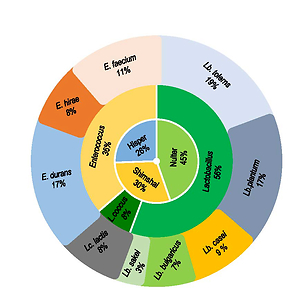
biochemical properties. The abundance of genera from the highest to the lowest
was lactobacilli (56%) followed by enterococci (36%) and lactococci (8%). Among
these isolates, 9 different species were recognized by studying their biochemical
characterization with the use of API 50 CHL systems. The dominance of species
included Lactobacillus paracasei subsp. tolerans (19%) followed by Lactobacillus
plantarum and Enterococcus durans (17%), Enterococcus faecium (11%),
Lactobacillus casei (9%), Enterococcus hirae and Lactococcus lactis (8%),
Lactobacillus delbrueckii subsp. bulgaricus (7%) and Lactobacillus sakei (3%).
Identification of 30 representative strains was further confirmed by 16S rRNA
gene sequence analysis. So, a foundation for developing local starter cultures of
LAB for the production of fermented products from yak milk is provided.
ACKNOWLEDGEMENTS
The authors are thankful for the financial
support of the Higher Education Commission of
Pakistan for the current research project under the
HEC Indigenous 5000 PhD Fellowship Programme
Phase II, Batch-I (PIN: 112-21766-2AVI-336).
CONFLICT OF INTEREST
The authors declare that there is no conflict of
interest.
REFERENCES (31)
1.
Abdelgadir W.S., Hamad S.H., Møller P.L., Jakobsen M., 2001.Characterisation of the dominant microbiota of Sudanese fermented milk Rob. Int. Dairy J. 11, 63–70, https://doi.org/10.1016/S0958-....
2.
Airidengcaicike, Chen X., Du X., Wang W., Zhang J., Sun Z., Liu W., Li L., Sun T., Zhang H., 2010. Isolation and identification of cultivable lactic acid bacteria in traditional fermented milk of Tibet in China. Int. J. Dairy Technol. 63, https://doi.org/10.1111/j.1471....
3.
Ao X., Zhang X., Zhang X., Shi L., Zhao K., Yu J., Dong L., Cao Y., Cai Y., 2012. Identification of lactic acid bacteria in traditional fermented yak milk and evaluation of their application in fermented milk products. J. Dairy Sci. 95, 1073–1084, https://doi.org/10.3168/jds.20....
4.
Bao Q.H., Liu W., Yu J. et al., 2012. Isolation and identification of cultivable lactic acid bacteria in traditional yak milk products of Gansu Province in China. J. Gen. Appl. Microbiol. 58, 95–105, https://doi.org/10.2323/jgam.5....
5.
Blaimont B., Charlier J., Wauters G., 1995. Comparative distribution of Enterococcus species in faeces and clinical samples. Microb. Ecol. Health Dis. 8, 87–92, https://doi.org/10.3109/089106....
6.
Chammas G.I., Saliba R., Corrieu G., Béal C., 2006. Characterisation of lactic acid bacteria isolated from fermented milk “laban”. Int. J. Food Microbiol. 110, 52–61, https://doi.org/10.1016/j.ijfo....
7.
Coeuret V., Dubernet S., Bernardeau M., Gueguen M., Vernoux J.P., 2003. Isolation, characterization and identification of lactobacillifocusing mainly on cheeses and other dairy products. Lait 83, 269–306, https://doi.org/10.1051/lait:2....
8.
Dewan S., Tamang J.P., 2007. Dominant lactic acid bacteria and their technological properties isolated from the Himalayan ethnic fermented milk products. Antonie Van Leeuwenhoek 92, 343–352, https://doi.org/10.1007/s10482....
9.
Gardiner G.E., Ross R.P., Wallace J.M., Scanlan F.P., Jägers P.P., Fitzgerald G.F., Collins J.K., Stanton C., 1999. Influence of a probiotic adjunct culture of Enterococcus faeciumon the quality of cheddar cheese. J. Agric. Food Chem. 47, 4907–4916, https://doi.org/10.1021/jf9902....
10.
Gran H.M., Gadaga H.T., Narvhus J.A., 2003. Utilization of various starter cultures in the production of Amasi, a Zimbabwean naturally fermented raw milk product. Int. J. Food Microbiol. 88, 19–28, https://doi.org/10.1016/S0168-....
11.
Hall T., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 41, 95–98.
12.
Hayes J.R., English L.L., Carter P.J., Proescholdt T., Lee K.Y., Wagner D.D., White D.G., 2003. Prevalence and antimicrobial resistance of Enterococcus species isolated from retail meats. Appl. Environ. Microbiol. 69, 7153–7160, https://doi.org/10.1128/AEM.69....
13.
Jasra A.W., Hashmi M.M., Waqar K., Ali M., 2016. Traditional yak herding in high-altitude areas of Gilgit-Baltistan, Pakistan: transboundary and biodiversity conservation challenges. In: W. Ning, Y. Shaoliang, S. Joshi, N. Bisht (Editors). Yak on the Move; Transboundary Challenges and Opportunities for Yak Raising in a Changing Hindu Kush Himalayan Region. International Centre for Integrated Mountain Development (ICIMOD). Kathmandu (Nepal), pp. 41–51.
14.
Kandler O., Weiss N., 1986. Genus Lactobacillus. In: P.H.A. Sneath, N.S. Mair, H.E. Sharpe and J.G. Holt (Editors). Bergey’s Manual of Systematic Bacteriology, Volume 2, 9th Edition. Williams and Wilkins. Baltimore, MD (USA), pp. 1063–1065.
15.
Liu W., Bao Q., Jirimutuet et al., 2012. Isolation and identification of lactic acid bacteria from Tarag in Eastern Inner Mongolia of China by 16S rRNA sequences and DGGE analysis. Microbiol. Res. 167, 110–115, https://doi.org/10.1016/j.micr....
16.
Luming D., Ruijun L., Zhanhuan S., Changting W., Yuhai Y., Songhe X., 2008. Feeding behaviour of yaks on spring, transitional, summer and winter pasture in the alpine region of the Qinghai-Tibetan plateau. Appl. Anim. Behav. Sci. 111, 373–390, https://doi.org/10.1016/j.appl....
17.
Mahmood T., Masud T., Imran M., Ahmed I., Khalid N., 2013. Selection and characterization of probiotic culture of Streptococcus thermophillus from dahi. Int. J. Food Sci. Nutr. 64, 494–501, https://doi.org/10.3109/096374....
18.
Marroki A., Zúñiga M., Kihal M., Pérez-Martínez G., 2011. Characterization of Lactobacillus from Algerian goat’s milk based on phenotypic, 16S rDNA sequencing and their technological properties. Braz. J. Microbiol. 42, 158–171, https://doi.org/10.1590/S1517-....
19.
Mathara J.M., Schillinger U., Kutima P.M., Mbugua S.K., Holzapfel W.H., 2004. Isolation, identification and characterization of the dominant microorganisms of kule naoto: the Maasai traditional fermented milk in Kenya. Int. J. Food Microbiol. 94, 269–278, https://doi.org/10.1016/j.ijfo....
20.
Mishra V., Prasad D.N., 2005. Application of in vitro methods for selection of Lactobacillus casei strains as potential probiotics. Int. J. Food Microbiol.103,109–115, https://doi.org/10.1016/j.ijfo....
21.
Ogier J.-C., Serror P., 2008. Safety assessment of dairy microorganisms: The Enterococcus genus. Int. J. Food Microbiol. 126, 291–301, https://doi.org/10.1016/j.ijfo....
22.
Oladipo I.C., Sanni A., Swarnakar S., 2013. Phenotypic and genomic characterization of Enterococcus species from some Nigerian fermented foods. Food Biotechnology. 27, 39–53, https://doi.org/10.1080/089054....
23.
Park J.-M., Shin J.-H., Lee D.-W., Song J.-C., Suh H.-J., Chang U.-J., Kim J.-M., 2010. Identification of the lactic acid bacteria in Kimchi according to initial and over-ripened fermentation using PCR and 16S rRNA gene sequence analysis. Food Sci. Biotechnol. 19, 541–546, https://doi.org/10.1007/s10068....
24.
Ren Y., Yang Y., Zhang D., Wang D., Zhang H., Liu W., 2017. Diversity analysis and quantification of lactic acid bacteria in traditionally fermented yaks’ milk products from Tibet. Food Biotechnol. 31, 1–19, https://doi.org/10.1080/089054....
25.
Sun Z., Chen X., Wang J. et al., 2011. Complete genome sequence of Lactobacillus delbrueckii subsp. bulgaricus strain ND02. J. Bacteriol. 193, 3426–3427, https://doi.org/10.1128/JB.050....
26.
Sun Z., Liu W., Gao W., Yeng M., Zhang J., Wu L., Wang J., Menghe B., Sun T., Zhang H., 2010. Identification and characterization of the dominant lactic acid bacteria from kurut: The naturally fermented yak milk in Qinghai, China. J. Gen. Appl. Microbiol. 56, 1–10, https://doi.org/10.2323/jgam.5....
27.
Tamura K., Stecher G., Peterson D., Filipinski A., Kumar S., 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6. Mol. Biol. Evol. 30, 2725–2729, https://doi.org.10.1093/molbev...
28.
Van de Casteele S., Vanheuverzwijn T., Ruyssen T., Van Assche P., Swings J., Huys G., 2006. Evaluation of culture media for selective enumeration of probiotic strains of lactobacilli and bifidobacteria in combination with yoghurt or cheese starters. Int. Dairy J. 16, 1470–1476, https://doi.org/10.1016/j.idai....
29.
Zagorec M., Champomier-Vergès M.-C., 2017. Lactobacillus sakei: a starter for sausage fermentation, a protective culture for meat products. Microorganisms 5, 56, https://doi.org/10.3390/microo....
30.
Zamfir M., Vancanneyt M., Makras L., Vaningelgem F., Lefebvre K., Pot B., Swings J., De-Vuyst L., 2006. Biodiversity of lactic acid bacteria in Romanian dairy products. Sys. Appl. Microbiol. 29, 487–495, https://doi.org/10.1016/j.syap....
31.
Zhang L., Yu Q., Han L., Zhang M., Yang L., Li Y.P., 2011. Evaluation of the potential probiotic properties of Lactobacillus strains isolated from traditional fermented yak milk. J. Food Agric. Environ. 9, 18–26.
CITATIONS (4):
1.
Single Molecule Real-Time Sequencing and Traditional Cultivation Techniques Reveal Complex Community Structures and Regional Variations of Psychrotrophic Bacteria in Raw Milk
Bingyao Du, Lu Meng, Huimin Liu, Nan Zheng, Yangdong Zhang, Shengguo Zhao, Jiaqi Wang
Frontiers in Microbiology
Bingyao Du, Lu Meng, Huimin Liu, Nan Zheng, Yangdong Zhang, Shengguo Zhao, Jiaqi Wang
Frontiers in Microbiology
2.
Leading report of molecular prevalence of tick borne Anaplasma marginale and Theileria ovis in yaks (Bos grunniens) from Pakistan
Muqaddas Nawaz, Rehmat Ullah, Zia Ur Rehman, Muhammad Naeem, Afshan Khan, Mohammed Bourhia, Muhammad Mudassir Sohail, Takbir Ali, Adil Khan, Tanveer Hussain, Furhan Iqbal
Archives of Microbiology
Muqaddas Nawaz, Rehmat Ullah, Zia Ur Rehman, Muhammad Naeem, Afshan Khan, Mohammed Bourhia, Muhammad Mudassir Sohail, Takbir Ali, Adil Khan, Tanveer Hussain, Furhan Iqbal
Archives of Microbiology
3.
Enhancing the application of probiotics in probiotic food products from the perspective of improving stress resistance by regulating cell physiological function: A review
Dingkang Wang, Ruijie Xu, Sha Liu, Xiaomin Sun, Tianxiao Zhang, Lin Shi, Youfa Wang
Food Research International
Dingkang Wang, Ruijie Xu, Sha Liu, Xiaomin Sun, Tianxiao Zhang, Lin Shi, Youfa Wang
Food Research International
4.
Isolation of Enterococcus hirae From Fresh White Yak Milk in Ledu District, Qinghai Province, China: A Comparative Genomic Analysis
Huimin Lv, Jiaqi Sun, Yuanyuan Guo, Guoxuan Hang, Qiong Wu, Zhihong Sun, Heping Zhang
Current Microbiology
Huimin Lv, Jiaqi Sun, Yuanyuan Guo, Guoxuan Hang, Qiong Wu, Zhihong Sun, Heping Zhang
Current Microbiology
Share
RELATED ARTICLE
We process personal data collected when visiting the website. The function of obtaining information about users and their behavior is carried out by voluntarily entered information in forms and saving cookies in end devices. Data, including cookies, are used to provide services, improve the user experience and to analyze the traffic in accordance with the Privacy policy. Data are also collected and processed by Google Analytics tool (more).
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.
You can change cookies settings in your browser. Restricted use of cookies in the browser configuration may affect some functionalities of the website.